Patterning cells over time: an added dimension in development
The differentiation of cells is a fundamental aspect of developmental biology. Scientists from the lab of Luisa Cochella have now found a novel mechanism that steers cell differentiation: patterns created not through spatial overlap of transcription factors, but through the combined action of transcription factors expressed at different times – even spanning multiple cell divisions. The findings are published in the current issue of the journal Developmental Cell.
Cells are the building blocks for tissues, organs, and ultimately complex organisms. As such, cells differ greatly in their structure, allowing them to fulfil different functions. Extreme examples are a nerve cell with a metre-long extension linking the spinal cord of a large mammal with a muscle, which in turn is composed of cells packed with crystal-like arrays of contractile proteins.
The mechanisms trough which cells differentiate into such specialists is one of the great themes of developmental biology. A “textbook mechanism” relies on proteins that control the activity of genes, so-called transcription factors. Transcription factors rarely work alone. Often, two or more transcription factors are expressed in different, but partly overlapping regions of an embryo; those cells where expression of both transcription factors coincide adopt a unique identity. This is called “spatial intersection” of transcription factors.
More than just patterning cells in space, transcription factors trigger different developmental programmes. Because of this, as cells divide, their progenies become increasingly different, specifying distinct cell lineages that ultimately give rise to different cells. Scientists from the lab of Luisa Cochella at the Research Institute of Molecular Pathology (IMP), Vienna BioCenter, have now added a new dimension to transcription factor intersections: time.
Tracking cell lineages as they diversify
The animal that Luisa Cochella’s lab uses as a model is the tiny nematode worm C. elegans. Worms of this species are complex organisms with a range of different tissues and organs, but they are composed of only about 1000 cells. The lineage of each of these cells is known throughout development, from fertilised egg to adult animal. This makes developing C. elegans embryos an ideal model for investigating the mechanisms that drive cell differentiation.
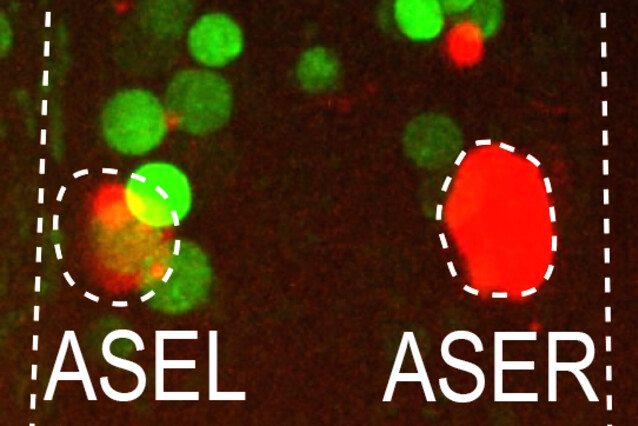
For years, Luisa Cochella and her colleagues have studied the role of lsy-6, a microRNA that can be found exclusively in the left one of two anatomically symmetric nerve cells, the ASE sensory neuron pair. Strikingly, the two cells also serve different functions, representing the only known directed asymmetry in the C. elegans nervous system to date. Asymmetric lsy-6 transcription requires the presence of the transcription factor CHE-1, which is present in both ASEs, where it is known to activate hundreds of symmetric genes. Therefore, the scientists knew that CHE-1 alone could not be responsible for the different lineage fates – the search was on for some intersectional patterning strategy.

The two lineage branches that give rise to the ASE neurons diverge at the 4-cell stage of an embryo, eight divisions before the rise of the ASE neurons themselves. The lineages are distinguished by the expression of two transcription factors (TBX-37 and TBX-38) in the precursor of the left ASE. The expression of these two transcription factors is required for the expression of lsy-6 four cell divisions later, when CHE-1 first appears. These were some of the clues that made Luisa Cochella think that TBX-37/38 would somehow act together with CHE-1 to activate lsy-6 specifically in the left ASE. However, TBX-37/38 were expressed in the lineage transiently, four cell divisions before CHE-1 and did not overlap spatially as in the known intersection mechanisms. How could these two transcriptional inputs work together to define a cell’s developmental fate without ever being together at the same time?
An experimental "tour de force"
To find out, Julien Charest and Thomas Daniele, a PhD student and a postdoc in the lab, teamed up with other lab members to establish a number of state-of-the-art genetics and genomics tools to study the lsy-6 regulation over time. First, they made sure that the function of TBX-37/38 was required only early and that these transcription factors did not stay around during the whole development. They then showed that TBX-37/38 bind directly to a sequence in the lsy-6 locus, and that this binding is necessary for the later activity of CHE-1 on lsy-6, demonstrating that there was a functional link between TBX-37/38 and CHE-1 despite their separation in time. In the right ASE neuron that doesn’t have early binding of TBX-37/38, CHE-1 cannot bind and activate lsy-6. Moreover, they could show that the function of TBX-37/38 is to induce antisense transcription of lsy-6 and this active transcriptional state is what primes lsy-6, making it competent for its later activation by CHE-1. Further experiments suggest that this priming mechanism is an epigenetic phenomenon that can be maintained in the absence of the original trigger, TBX-37/38. Understanding the basis of this mechanism ultimately allowed the researchers to identify other neuron pairs that also display asymmetry in gene expression caused by TBX-37/38 priming in the left members of these pairs.
“The spatial intersection mechanisms we knew all require that cooperating transcription factors must be present in the same cell, at the same time”, said Luisa Cochella. “We discovered a mechanism that is created by two transcription factors which act on the same gene but at different times – separated by four cell divisions. We called this a ‘temporal intersection’.”
The scientists believe that temporal intersections could be a common mechanism driving the generation of cell diversity during development. An implication of this mechanism is that the activity of a transcription factor on a given gene depends on the previous history of the cell and which other transcription factors acted on that gene earlier. As such, temporal intersections are important not only for understanding the fascinating process of development, but also for strategies that use transcription factors to differentiate cell types in culture.
Original Publication
Julien Charest, Thomas Daniele, Jingkui Wang, Aleksandr Bykov, Ariane Mandlbauer, Mila Asparuhova, Josef Röhsner, Paula Gutiérrez-Pérez and Luisa Cochella (2020): "Combinatorial action of temporally segregated transcription factors". Developmental Cell. DOI: https://doi.org/10.1016/j.devcel.2020.09.002
About the Vienna BioCenter PhD Programme
Much of the work underlying this publication was done by a doctoral student of the Vienna BioCenter PhD Programme. Are you interested in a world-class career in molecular biology? Find out more: https://training.vbc.ac.at/