Scientists show structure of enzyme involved in protein degradation
An international team of scientists including Zuzana Hodakova and David Haselbach used cryo-electron microscopy to analyse the structure of the enzyme UBR5. Difficult to work on due to its size, UBR5 is important due to its role in marking proteins for degradation. The findings were now published in EMBO Journal.
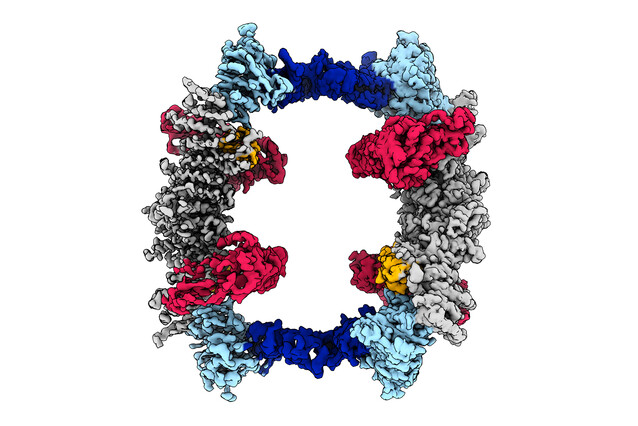
Structural biologists David Haselbach and Zuzana Hodakova are co-corresponding authors of a publication that presents the structure of UBR5, a large HECT-domain-containing ligase of the E3 class. The enzyme plays an important role in marking proteins for degradation. To learn more about the finding, we talked to David and Zuzana.
You and your collaborators report the previously unknown structure of a very large enzyme. Tell us a bit more about the background of the project.
Zuzana: There are not many known structures in this class of enzymes, HECT-E3 ligases. The only known structure from this class of three enzymes until now was HUWE1, which was solved by the Clausen lab in collaboration with us here at the IMP in 2021. We are dealing with a very heterogeneous class of E3 ligases here, and very little is known about them.
And why are not more of them known?
Zuzana: I think that it is just very difficult to work with them. We know that they are very large. They're single polypeptide enzymes and they are enormous; UBR5, for instance, is over 300 kilodalton large, and HUWE1 is even a bit larger. This is the fifth largest E3 ligase from this class with 28 members. Working out their structure is also hard because they contain multiple domains, many of them are very flexible - and simply difficult to handle for structural analysis.

How did you overcome this?
David Haselbach: I think first and foremost, it took some courage to tackle this. In the recent past, there were reports that you could do it, but most people shied away from trying because it is technically hard. I think the courage to just try to do it was an important first step.
How and when did your work on UBR5 start?
In our 2021 study on Akirin2, there was this side result of a CRISPR-screen that UBR5 plays a role in the degradation of myc and might play a role in the whole nuclear transport of the proteasome. A result that I always liked, and I wanted to address in more detail. But when we wrote the Akirin paper, it quickly got too large for side-results, and our collaborators in the Zuber lab also prioritised other aspects. So, at the time, we decided against further details on UBR5. This left looking at UBR5 beyond its nuclear transport role as an obvious next step.
Zuzana: The first time that we purified UBR5 as a “nice” protein that behaves was in March 2022. From there, it took us about two months until we had a first large and high-quality collection on the Krios [a high-end cryo-electron microscope] to actually start working on the structure. Which we did for about a year now.
How was your approach different from that of other labs working in similar topics?
David: There were in fact several labs working on related questions. Luckily for us, we didn't know most of the time that there were! I guess we would have given up from the start already, because the other labs had already started two or three years earlier. What I find fascinating is that the other labs all have different reasons for working on UBR5, they come from very different questions and went into different directions. One lab is the lab of Nico Thomä, who asked how certain receptors get degraded. And they found in their screens that this happens via UBR5. Another lab, the lab of Brenda Schulman, is more interested in how HECT E3 ligases work - and found specifically UBR5 as the most interesting one. But as Zuzana said, there are 28 of them, and there is no reason why UBR5 must be the next one to study. There was also a fourth lab that also has published now, which were the very first ones that we actually knew of, because they had a bioRxiv paper on the structure.
We were finishing up most of the story, and Zuzana submitted an abstract for a conference. Then we got approached by colleagues and they told us, hey, we are competing actually, maybe we should talk. This resulted in coordination of the publications and all of the stories being published.
From what we know from publications that are now on bioRxiv or by the talks that we have seen at conferences, larger labs put much more resources into the project. So there is a much larger group of people working on it, their workflows are more established – while we started our work quite naively.
Why are more people working on a project an advantage?
David: I think that there is a difference between our stories and their stories. Some of the other approaches originate from a different biological question, which then in the end resulted in also working on the UBR5 structure. For example, from the publication by Michael Rape and Nico Thomä, this was a merge collaboration where forces of two labs joint. One worked on the biological question, the other on the structural part, and then it came together.
What I think that gave us an advantage was that we were directly interested in the structural part. Alongside the expertise in our lab, I think that this focus is what really pushed the project. I think that we are very good when it comes to cryo-electron microscopy. But what really made the difference is that we just knew how to get from point A to point B. We had the expertise and then we just pushed to make it happen.
UBR5: the animated structure
We use a YouTube plugin to display social media content on this page, which places requests to YouTube servers. These requests make your IP address visible to YouTube, who may use it in accordance with their data privacy policy. Please agree to make the YouTube video visible. You can find more information in our privacy settings.
AcceptAnd now you have the structure. So what do you do with that? What's next?
Zuzana: Well, it's not only the structure. We also found a protein that interacts with UBR5, which was one of the things that started this project. It interacts with Akirin2, which is essential for the import of the proteasome into the nucleus. We are also trying to figure out the role between UBR5 and Akirin2 in this interaction: if it plays a role in the proteasome import or if it interacts, for example, with something already passed in the nucleus, the import, or some other mechanism. So now we're trying to learn about the functions of UBR5 in more detail.
Doing so, I'm going more into the into the waters of Hanna Brunner in the lab, who is working actively on the import of the proteasome into the nucleus. There is some overlap with her project now, which is very interesting. But even this structure, even this paper on UBR5, from the lab perspective, it was me and Irina Grishkovskaya.
David, could you describe the role of Zuzana in steering this project a bit more?
David: Zuzana drove the project very independently of me and of others, and I still think she knows much more about HECT E3 ligases than I do. There are 600 of E3 ligases, and of those only a small subset are HECT E3 Ligases. But they all lead to the degradation of important things. So there is a huge body of work that Zuzana quickly found out about.
She was really driving the project. What I did was mentoring and a little bit of steering. Zuzana asked the questions; she designed what to do next. She managed the people. She talked to the collaborators. She was running the project basically, while supervising herself somehow.
And how does the structure fit into the bigger picture of things now? What's the significance? Why is this important?
David: We still don't know how HECT E3 ligases work at all. What is the chemical mechanism? There is an interesting model, but probably wrong. Nevertheless, E3 ligases are heavy targets for drug discovery now. A lot of people try to do something in this direction. That may or may not be something for us to following up on.
On other end, the structure is definitely another puzzle piece in our endeavour to understand nuclear degradation. Unfortunately, nuclear degradation isn't really studied because the ubiquitin proteasome system is very well studied in the cytoplasm. Our previous paper already showed that nuclear degradation is really important, and we should seek to better understand how degradation in nucleus and cytoplasm differ. This is something we will definitely investigate further.
UBR5 is one of a few E3 ligases that are exclusively nuclear, at least in our current understanding. And that's quite something. I think one puzzle piece that I find fascinating, which is pretty much in the title of our paper, is that we find that UBR5 is a chain elongating the E3 ligase. So it's much faster, at least in our hand. It's much faster to put longer chains of ubiquitin on a protein rather than just initialize the first one. It might act as an associate enzyme that doesn't take the decision whether something is degraded, but instead makes degradation more efficient. And that's an idea that I feel can be followed up in the future.
And what was the role of your collaborators?
David: Right, there is a third co-corresponding author, Nick Brown from the University of North Carolina in the US. Nick has made major contributions: he's an expert in biochemistry and E3 mechanisms, and I think without him we wouldn't have managed much of it in this time. The background story is that both of us were already collaborating during our postdocs. He was in Brenda Schulman’s lab, and we decided after our postdocs to go on collaborating. And this proved very fruitful.
Both of us could use our strengths. And Zuzana also had mentoring meetings and many discussions with Nick. I think that helped us a lot, because we are really good with solving structures. But if there is someone who really knows the specific biochemistry of E3 biochemistry, that is definitely also a secret to success and Nick’s expertise. He sent us a lot of enzymes and chemicals that we needed for assays, and also protocols, how to do certain assays. If we would have needed to establish all essays and all the things without help on our own, we probably would have needed another year.
Zuzana: Sometimes there was a lot of stuff coming from Nick and David, but it was very nice how they mentored me and how they gave me the freedom to really try and do things on my own. I'm a postdoc, so the next possible career choice would be to try to do this on my own, right? And they were really showing me how an independent project works and how one must do these things.
Original Publication
Zuzana Hodáková, Irina Grishkovskaya, Hanna L. Brunner, Derek L. Bolhuis,
Katarina Belačić, Alexander Schleiffer, Harald Kotisch, Nicholas G. Brown, David Haselbach (2023): “Cryo-EM structure of the chain-elongating E3 ubiquitin ligase UBR5”. EMBO Journal. DOI: 10.15252/embj.2022113348
Further reading