Structure of transcription-export complex TREX revealed
TREX ("TRanscription-and-EXport complex") is a key player in the mRNA export pathway. In a new study published in eLife today, scientists from the lab of Clemens Plaschka report the three-dimensional structure of the human TREX core complex.
True to its name, messenger RNA (mRNA) conveys the instructions from our DNA genome to the cellular machinery that translates these into proteins. However, mRNA is produced in the cell’s nucleus, where the DNA is located, and must be transported into the cytoplasm before it can be translated. This poses a challenge: only a faithfully made mRNA, carrying all the necessary RNA modifications and partner proteins, should be allowed out of the nucleus. A key player in the mRNA export pathway is the transcription-and-export complex (TREX). In a new study published in eLife today, scientists from the lab of Clemens Plaschka report the three-dimensional structure of the human TREX core complex.
Ticking off the protein to-do list
Newly made mRNA undergoes several RNA modifications such as capping, splicing, and cleavage and polyadenylation. Like checkmarks on a to-do list, proteins bind the mRNA after each modification step to mark its successful completion. The TREX complex then binds the modified mRNA-protein assembly to license it for export into the cytoplasm. “However, little is currently known about the mechanisms and structural basis of how mRNA packaging and export licensing occur”, explains Plaschka. “TREX binds several of these checkpoint marks on the mature mRNA and prepares it for export, yet how can a single complex select the mature mRNA and license it efficiently?”
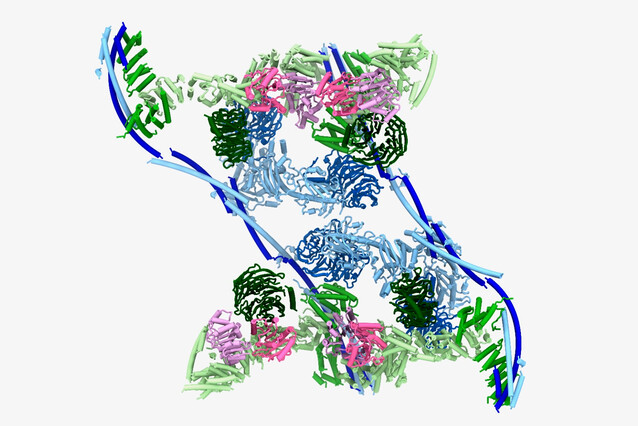
To shed light on this process, Plaschka and his group, including Master student Thomas Pühringer and Postdoc Ulrich Hohmann, who is jointly affiliated with the lab of Julius Brennecke (Institute of Molecular Biotechnology, also at the Vienna BioCenter), used cryo-electron microscopy to determine the three-dimensional structure of the TREX complex at 3.3 Ångström resolution – with a surprise. “We had assumed that the human TREX core would be a complex of seven different proteins, each in a single copy. To our astonishment, we instead found that TREX forms a large multimer: it assembles into a 28-subunit tetramer, with each protein present in four copies.”
Multiplicity increases efficiency
This unexpected organisation can explain how TREX can simultaneously bind several “checkmarks” on the maturing mRNA. “The tetrameric TREX complex contains multiple binding sites for maturation marks. Once one of the TREX monomers has bound one mark, the remaining three monomers would be brought into close proximity of the mRNA, making the recognition of subsequent marks more likely. Similarly, this can also explain how the subsequent licensing of mRNA for export is made more efficient, by providing multiple binding sites for loading the so-called mRNA export factor onto mRNA.”
The structure also sheds light on human disorders. “We could map several mutations linked to diseases onto our three-dimensional model, informing us on how some mutations disrupt the structure and how potential consequences may arise.”
The exciting path ahead
For Plaschka, the eLife study is exciting also on a personal level. “This is the first paper out of my lab, which we started at IMP in 2018. Going from an empty lab space to our first paper was a challenging but exciting process, made possible through the great and combined efforts of all our group members and the outstanding support junior group leaders receive at the IMP at multiple levels.”
Together with the ERC Starting Grant, which Plaschka was awarded in 2020 for work on mRNA packaging and export, this study is the starting point for a new research program that will probe the final stages of mRNA production in the nucleus. “mRNA is transcribed, processed, packaged and exported. Beyond the current study, we are excited to better understand the bridges between these steps and investigate how they are coordinated, regulated, and integrated.” Plaschka is looking forward to recruiting more researchers to his group. “We are now expanding projects based on this first structure and offer exciting opportunities for Master, PhD, and Postdoctoral researchers.”
Original Publication
Thomas Pühringer, Ulrich Hohmann, Laura Fin, Belén Pacheco-Fiallos, Ulla Schellhaas, Julius Brennecke, Clemens Plaschka: "Structure of the human core transcription-export complex reveals a hub for multivalent interactions". eLife 2020;9:e61503 DOI: 10.7554/eLife.61503
About the Vienna BioCenter PhD Programme
Like all research groups of the IMP, the Plaschka Lab participates in Vienna BioCenter PhD Program. Are you interested in a world-class career in molecular biology? Find out more: https://training.vbc.ac.at/