Tracking cells in the regenerating limb through space and time
In order to understand how tissues arise during embryonic development and regeneration, scientists need to find a way to follow different cell lineages in space and time as they divide and migrate. Tzi-Yang Lin, PhD student in the lab of Elly Tanaka, has developed a transgenic frog line to do just that, marking specific cells with a fluorescent protein and tracking their fate. He tells us more about his work, now published in the journal Development, Growth, & Differentiation.
Tzi-Yang, you developed a new transgenic line of the widely used model frog, Xenopus laevis. How does one do that?
The method is called restriction enzyme-mediated integration. You start by collecting sperm cells from a male frog and then extract their nuclei. You mix these nuclei with many copies of a circular piece of DNA, a plasmid, that carries the genes you want to integrate into the sperm genome. After that, you add a restriction enzyme, which is comparable to a pair of scissors that cuts the DNA at multiple locations. Through a natural repair mechanism, the plasmid DNA is integrated into the sperm nuclear DNA. Once this has worked, you can collect these nuclei and inject them into frog eggs, which will then be fertilised and develop into transgenic tadpoles that carry the genes you wanted to use. The concept is quite simple, but it’s actually a complicated process that takes a very long time.
What kind of line did you work on?
I engineered this line so that specific cells in the arms and legs could be labelled with a fluorescent protein. The frogs I used produce the green fluorescent protein, or GFP, in all tissues, such that the whole animal glows green. The GFP gene sits between two copies of a short DNA sequence that can be recognised and cut by an enzyme, the Cre recombinase. However, this enzyme can only be produced if the gene that encodes it is activated by a promoter. In this case, I made it so that the promoter is only working in the cells that I wanted to label. If the promoter is active, the enzyme is produced and cuts out the GFP gene, allowing the next gene – which encodes a magenta marker – to be expressed. This magenta marker cannot be expressed as long as the GFP gene is there. The result is a green, fluorescent frog, with only a handful of cells marked in magenta where the tissue-specific promoter is active.
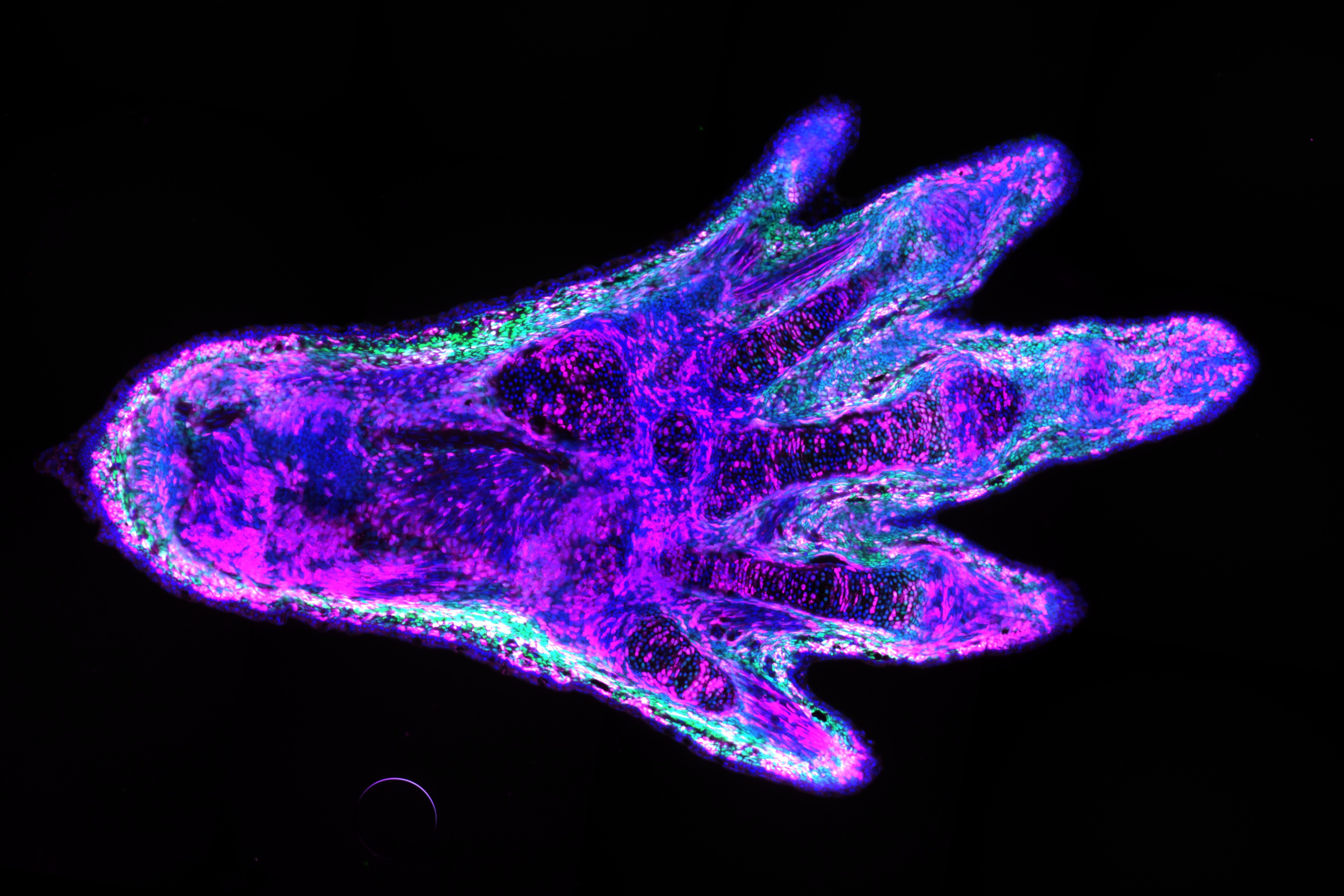
Why do you need to mark specific cells?
In the lab of Elly Tanaka, our ultimate goal is to understand how amphibian limbs can regrow after they were amputated. An important aspect is to know which cell types contribute to the regeneration process. Tracking specific cells will allow us to understand which ones are crucial to the formation of each tissue in the newly formed limb. The ability to label and isolate specific cell types also opens the door to study the molecular mechanisms of limb regeneration at a cell type-specific level. This was demonstrated by a previous paper in our lab, where my colleagues studied the contribution and the genetic programs of the limb fibroblasts during the limb regeneration of axolotls.
Xenopus frogs can partially regenerate their limbs, but don’t regrow fully functional legs like salamanders do, so we’re interested in labelling and tracing frog cells during regeneration to see what the differences are. We previously published a frog transgenic line that allows tissue-specific cell labelling and tracing. In the current methods paper, I detailed the process of the technology development and characterised our transgenic lines. In this case, I labelled fibroblasts, but in theory, the method could be applied to other cell types that have tissue-specific promoters.
Have other scientists tried to label specific cells in frogs before?
Yes, the methods to genetically label specific cell types with fluorescent proteins have been used in frog research. It’s usually done by expressing fluorescent proteins under the control of tissue-specific promoters. However, these methods are not suitable for cell tracking. In order to track the marked cells, the cells have to be permanently labelled. On top of that, it requires an inducible system that only labels cells at a specific time. If this were not the case, we wouldn’t be able to trace cells because of the inability to distinguish the newly labelled cells from the old ones. This strategy has been widely used in mouse research, but it hadn’t been successfully demonstrated in frogs until now.
Original publication
Tzi-Yang Lin, Yuka Taniguchi-Sugiura, Prayag Murawala, Sarah Hermann, Elly M. Tanaka: “Inducible and Tissue-Specific Cell Labelling in Cre-ERT2 Transgenic Xenopus lines”. Development, Growth, & Differentiation (2022). DOI: 10.1111/dgd.12791.
Further reading
