In situ structural biology and transposable elements
Sydney Brenner once famously stated, albeit provocatively, that "progress in science depends on new techniques, new discoveries, and new ideas, probably in that order". In that spirit, we combine technical development in cryo-focused ion beam milling (cryo-FIB) and cryo-electron tomography (cryo-ET) with biological applications to address questions in cellular structural biology in our group. Using cryo-FIB milling, we generate thin sections of biological samples that are subsequently imaged in a transmission electron microscope at different tilt angles. The resulting projections of the cellular material can be reconstructed similar to computed tomography widespread in biomedical applications. These approaches allow to generate three dimensional reconstructions of the cellular environment that reveal membrane architecture and visualise single protein complexes. Through computational analyses, the protein complex identified in these tomograms can be extracted, averaged, and, in favourable cases, even resolved to high resolutions below 1 nanometre, revealing secondary structure or even side-chains.
These technologies recently allowed us to study the cellular structural biology of a transposon or "jumping gene", the copia retrotransposon.
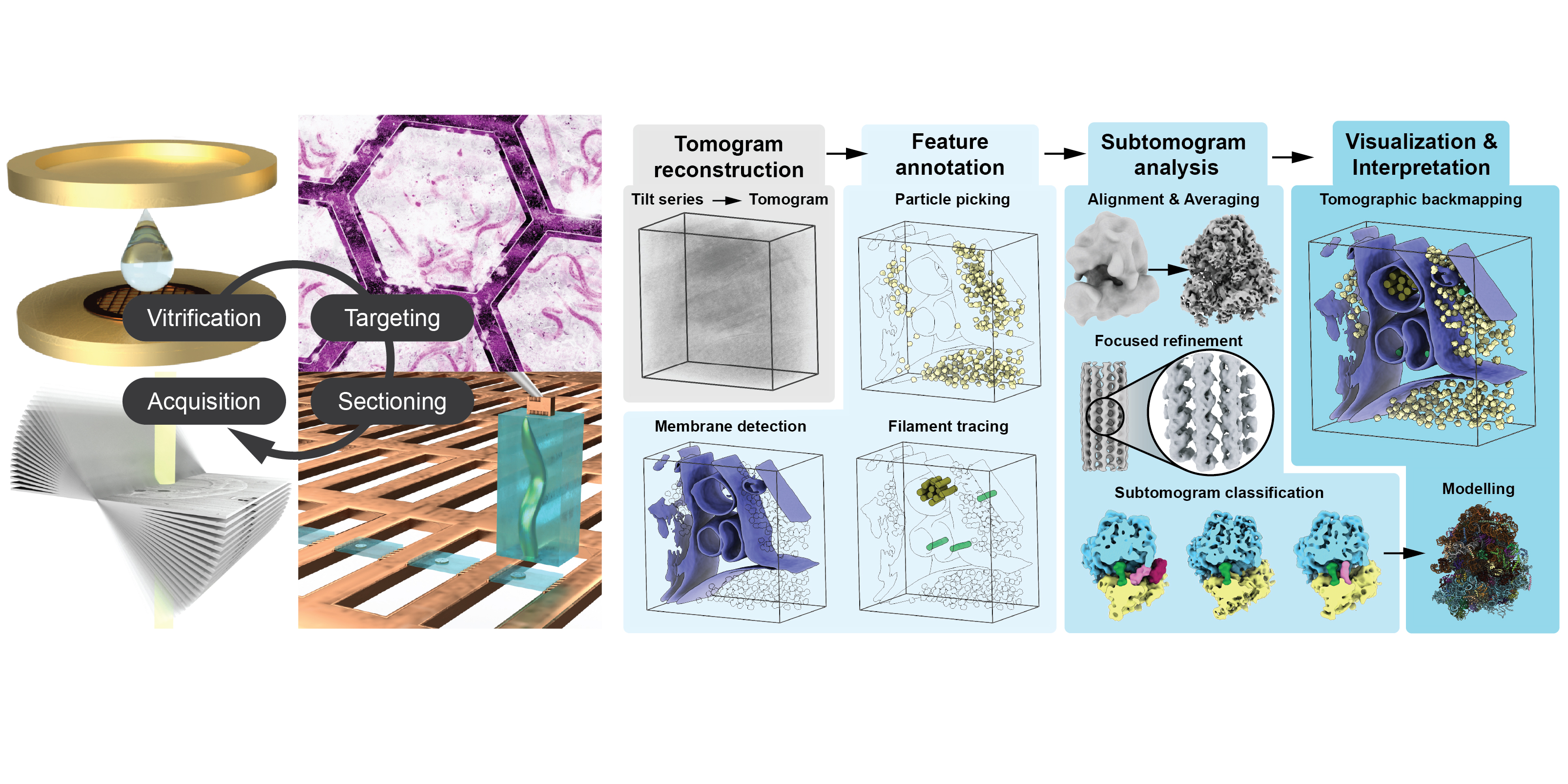
Transposable elements (TEs) are a diverse group of mobile genetic elements that can autonomously replicate and insert themselves into new genomic locations. TEs are widespread in organisms across the tree of life, frequently constituting a substantial proportion of their host genomes. In mammals, for instance, TEs account for approximately 25-75% of the genome, with humans reaching ~50%, and in some plant species, TEs can comprise up to 85% of the genome. A subclass of TEs, the long terminal repeat (LTR) retrotransposons, of which the copia retrotransposon is a member, transpose via an RNA intermediate and are evolutionarily related to retroviruses. If not kept in check, TEs pose a severe threat to genome integrity and intricate defence mechanisms have evolved to protect metazoan germline genomes from this imminent threat by transcriptional and post-transcriptional regulation. Currently, we are fascinated by the cell biological mechanisms that drive the replication cycle and, thus, evolutionary success of TEs. How do retrotransposons reach the germline genome to generate new insertions? What drives the sex-specificity observed for TEs? And how do TEs make their way into the genome in the first place? To that end, we use the model system Drosophila melanogaster as well as other model systems to study TEs in their respective niches during gametogenesis, the generation of sperm and egg.
Fig. 2: Video illustrating the visualisation of copia retrotransposons within fruit fly (D. melanogaster) egg chambers. For details, please see: doi.org/10.1016/j.cell.2025.02.003
In addition to these specific biological questions, we thrive to make new discoveries by developing and improving methodology, hardware, and open-source software in cryo-FIB milling to enable new experimental approaches and widen the sample spectrum in cryo-ET. Recent work in the field by us and others has opened up the world of tissue biology to the cellular structural biologist. By employing micromanipulation methods to extract material from large vitreously frozen multicellular organisms and tissues, a process referred to as lift-out, we are able to zoom into cellular ultrastructure of tissues at molecular resolution.
Fig. 3: Video illustrating the process of manipulation of a biological sample (here a C. elegans larva) inside the FIB-SEM instrument, referred to as cryo-lift-out. This methodology renders multicellular organisms and tissues amenable to high-resolution visualisation by cryo-ET. For more details, see: https://doi.org/10.1038/s41592-023-02113-5
Moving forward, we aim to leverage our developed approaches to investigate the cellular structural biology of germ cells, with a particular focus on the architecture and interactions of LTR retrotransposons within them. By preserving native cellular environments at near-atomic resolution, these techniques will enable us to uncover key structural insights into retrotransposon activity, regulation, and their impact on germline integrity.
Selected Publications
- Klumpe S., Senti K.A., Beck F., Sachweh J., Hampoelz B., Ronchi P., Yeroslaviz A., Briggs J.A.G., Brennecke J., Beck M., Plitzko J.M. (2024). In-cell structure and snapshots of copia retrotransposons in intact tissue by cryo-electron tomography. bioRxiv (DOI: 10.1101/2024.02.21.581285), Cell (DOI: 10.1016/j.cell.2025.02.003).
- McCafferty C.L., Klumpe S., Amaro R.E., Kukulski W., Collinson L., Engel B.D. (2024). Integrating cellular electron microscopy with multimodal data to explore biology across space and time. Cell 187(3): 563–584 (DOI: 10.1016/j.cell.2024.01.005).
- Schioetz O.S., Kaiser C.J.O., Klumpe S., Morado D.R., Poege M., Schneider J., Beck F., Thompson C., Plitzko J.M. (2023). Serial Lift-Out – Sampling Molecular Anatomy in Whole Organisms. Nature Methods (accepted manuscript).
- Klumpe S., Fung, H.K.H., Goetz S.K., Zagoriy I., Hampoelz B., Xiaojie Z., Erdmann P.S., Baumbach J., Müller C.W., Beck M., Plitzko J.M., Mahamid J. (2021). A Modular Platform for Streamlining Automated Cryo-FIB Workflows. eLife 10:e70506 (DOI:10.7554/eLife.70506).
- Schioetz O.S., Klumpe S., Plitzko J.M., Kaiser C.J.O (2024). Cryo-electron tomography: en route to the molecular anatomy of organisms and tissues. Biochemical Society Transactions (BST20240173).
Join us
- Master students and Post-docs: Contact Sven Klumpe with a letter of intent detailing why you want to join the lab.
- PhD students: Calls open 1 March and 1 September, apply here: